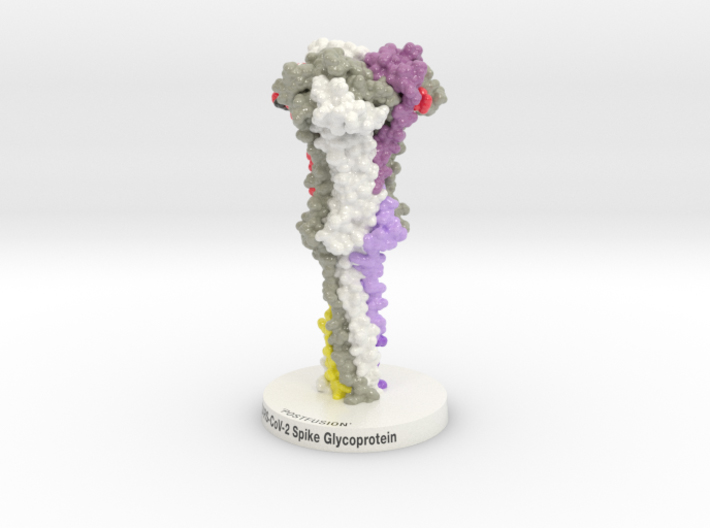
The outbreak of a novel coronavirus (SARS-CoV-2) represents a pandemic threat that has been declared a public health emergency of international concern. The SARS-CoV-2 Spike Glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics. Biologic Models designed this 3D printed protein model of the SARS-CoV-2 Spike Glycoprotein Postfusion 6XRA to help scientists better visualize these targets.
Biologic Model of SARS-CoV-2 Spike Glycoprotein in its Postfusion or Inactive State and 3D printed and colored-coded to denote functional domains (NTD blue, RBD green, SD1, light tan, SD2 red-orange-yellow, FP cyan, RRAR brown). After cleavage at the S2′ site, (dark grey), Spike undergoes significant structural rearrangement extending temporarily out from its central core to an elongated Intermediate Fusion state before recoiling into the structure seen here.
Biologic Explorer: 6XRA
To better understand the structural relationship of the domains within the Spike Glycoprotein, a protein dataset 6XR8 has been colorized to match our 3d printed model. Explore each domain below after “Initializing” the dataset.
SARS-CoV-2 Spike Glycoprotein
The membrane-bound configuration of SARS-CoV-2 Spike Glycoprotein (PDB ID: 6XRA seen below) is a trimer complex. Each monomer of the trimer is broken into two primary domains (S1 and S2) and then further characterized by a series of functional domains that facilitate either host identification and membrane fusion.
SCIENCE Image Gallery
Text and Illustrations from SCIENCE publication: “Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation.”
At the beginning of each S1 domain of the Spike Glycoprotein is the N-terminal Domain (Blue). It is associated with the stabilization of adjacent monomers of the Spike Glycoprotein. Following it is the Receptor-Binding Domain (RBD green) that binds angiotensin-converting enzyme 2 (ACE2) with a higher affinity than it does severe acute respiratory syndrome SARS-Coronavirus ‘Classic’ (2002). This in part is due to the unique RBD Binding Motif (dark green) that matches the binding motif of a similar Coronavirus found in Pangolins. Several published SARS-CoV-1 RBD-specific monoclonal antibodies were tested and found that they do not have appreciable binding to SARS-CoV-2, suggesting that antibody cross-reactivity may be limited, in part due to changes to these residues (Dark Green).
Spike protein S2 (residue 686-1273) mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. During viral and target cell membrane fusion, the coiled-coil regions (light yellow and orange) assume a trimer-of-hairpins structure, positioning the Fusion Peptide (cyan) in close proximity to the C-terminal region of the ectodomain. The formation of this structure appears to drive the apposition and subsequent fusion of viral and target cell membranes.
Model Description
Biologic Model of SARS-CoV-2 Spike Glycoprotein Postfusion 6XRA 3D printed in Full-color Sandstone or Plastic and colored to match “Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation” published recently in SCIENCE.


