SARS-CoV-2 Structural Proteins
The outbreak of a novel coronavirus (SARS-CoV-2) represents a pandemic threat that has been declared a public health emergency of international concern. The structural proteins of the SARS-CoV-2 Virion are key targets for vaccines, therapeutic antibodies, and diagnostics. Biologic Model designed the 3D printed SARS-CoV-2 Virion Model to help scientists better visualize the structural assembly of viral proteins. This scientific model is being used to help facilitate the rapid development and evaluation of medical countermeasures to address the ongoing public health crisis.
SARS-CoV-2 Structural Proteins
The structural proteins that define the SARS-CoV-2 Virion are composed of the Envelope Protein (E Protein tined cyan), the Membrane Protein (M Protein tinted green), the Nucleocapsid Protein (N Protein tinted blue), and the Spike Glycoprotein (S Protein tinted yellow).
3D Print SARS-CoV-2 Virion
Model Description
Biologic Model of SARS-CoV-2 Virion 3D printed in Multi-color Transparent Plastic. SARS-CoV-2 Virion is 3D printed and colored-coded to denote structural proteins by color (Spike Protein: Yellow, Membrane Protein: Cyan, Envelope Protein: Green, and Nuclear Protein: Blue) and tinted from N to C terminal darkest to lightest.
Video
SARS-CoV-2 Spike Glycoprotein
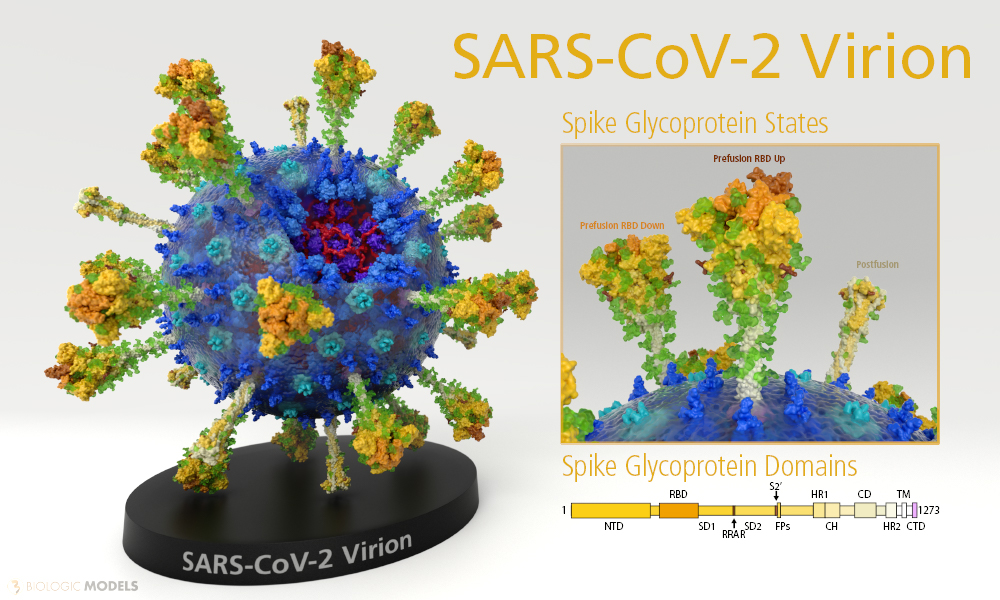
The nCoV Spike (S) Glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics. The predominant state of the Spike is a trimer complex that locates the three receptor-binding domains (Green) after a hinge region. The RBD in its ‘Down’ position can be seen above. In its ‘Up’ position, the RBD rotates up making the binding residues of the Virus accessible to ‘Hook’ onto the Human ACE Receptor. Several published SARS-CoV RBD-specific monoclonal antibodies were tested and found that they do not have appreciable binding to 2019-nCoV S, suggesting that antibody cross-reactivity may be limited between the two RBDs. The structure of SARS-CoV-2 Spike Glycoprotein 6VXX should enable the rapid development and evaluation of medical countermeasures to address the ongoing public health crisis.
The membrane-bound configuration of SARS-CoV-2 Spike Glycoprotein is a trimer complex. Each monomer is composed of a series of subdomains that play functional roles in either host attachment or membrane fusion.
During infection, the Spike Protein’s S1 domain attaches the virion to the cell membrane by interacting with host receptor. This initiates infection and subsequent membrane fusion. Binding to human ACE2 receptor induces conformational changes in the S glycoprotein. Proteolysis of the fusion peptide (white) of S2 domain activates membrane fusion within endosomes. The Spike protein S2 domain (light tan) mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. (See another example of a viral Fusion Glycoprotein LINK).
To learn more about the SARS-CoV-2 Spike Glycoprotein, read our in-depth investigation of its conformation changes: Read More
Biologic Explorer: SARS-CoV-2 Spike Glycoprotein 6VXX
Explore the SARS-CoV-2 Spike Glycoprotein in its “Down/Closed” configuration. 3D model constructed from PDB ID: 6VXX
3D Print SARS-CoV-2 Spike Glycoproteins
Healthcare providers use our 3D printed scientific models of the SARS-CoV-2 Spike Glycoprotein to discuss topics like the mRNA vaccine or how antibodies block the virus from infecting cells. Available in both Full-color Sandstone and Plastic.
SARS-CoV-2 Envelope Protein
The Envelope Protein (E Protein) plays a central role in virus morphogenesis and assembly. The E Protein self-assembles in host membranes forming pentameric protein-lipid pores. Each E Protein Complex functions as a mode of ion transport. They also play a role in the induction of apoptosis activating the host NLRP3 inflammasome, causing to IL-1beta overproduction and eventual cell death.
Biologic Explorer: SARS-CoV-2 Envelope Protein 5X29
While a complete dataset of SARS-CoV-2 Envelope Protein has yet to be crystallized as of the date of this posting, the SARS-CoV-1 Envelope Protein is available to explore. A homology model it was used to create our SARS-CoV-2 Virion Model. Explore protein dataset 5X29 below.
SARS-CoV-2 Membrane Protein
Component of the viral envelope that plays a central role in virus morphogenesis and assembly via its interactions with other viral proteins. The Membrane protein (M Protein) is a transmembrane protein that stabilizes the Nucleoprotein to the plasma membrane. CRYO-EM ultrastructure analysis of SARS-CoV-2 reveals M protein interacts with both the S Protein and N Proteins providing scaffolding for viral assembly.
While a complete dataset of SARS-CoV-2 Membrane Protein has yet to be crystallized as of the date of this posting, a collection of datasets closely approximate subdomains of the Membrane Protein. Explore homology models 5CTG (N-terminal), 6QPK, and 4O7J (C-terminal Domain) below.
Biologic Explorer: SARS-CoV-2 Membrane Protein 5CTG, 6QPK, 4O7J
SARS-CoV-2 Nucleoprotein
The Nucleoprotein (N Protein) packages the positive strand viral genome RNA into a helical ribonucleocapsid (RNP) complex. The N Protein plays a fundamental role during virion assembly through its interactions with the viral genome and membrane protein. It also plays an important role in the enhanced efficiency of viral RNA transcription.
While a complete dataset of SARS-CoV-2 Nucleoprotein has yet to be crystallized as of the date of this posting, a collection of subdomains have. Explore datasets 6VYO (N-terminal RNA-binding domain), and 7C22 (C-terminal Domain) below.