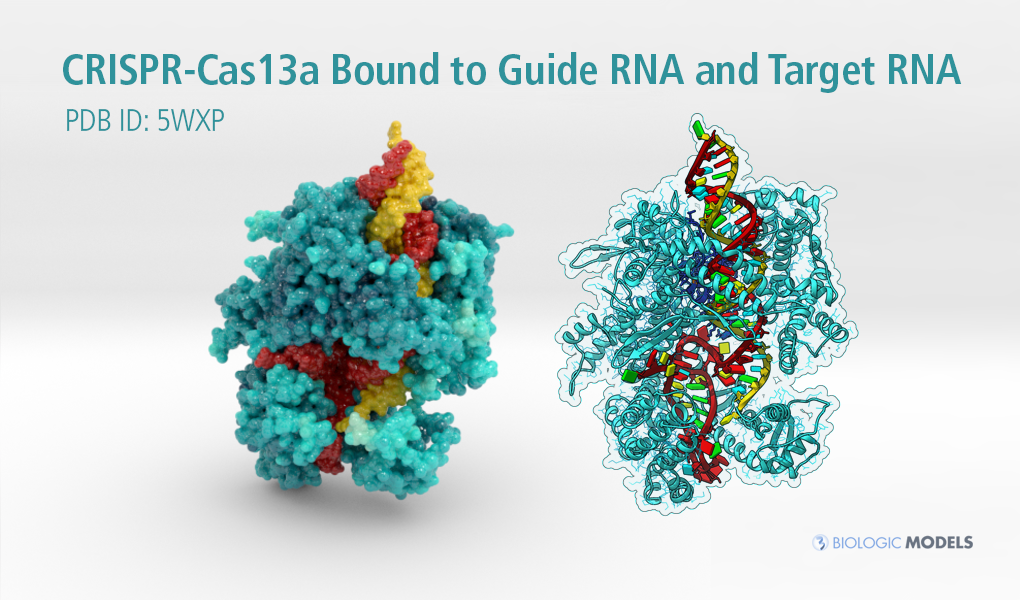
CRISPR-Cas13a Bound to Guide RNA and Target RNA
CRISPR-Cas13a is a newly discovered member of the Genome editing system making waves in the biotech community. Explore this 3D printed protein model of CRISPR-Cas13a to examine how it uses RNA to target and edit DNA. Protein model created from PDB ID: 5XWP
Protein Description
CRISPR is a family of genome editing proteins that evolved in bacteria and archaea to combat viral infections. CRISPR captures portions of infected viral DNA and cuts out specific portions to save as a guide to find viral DNA should the virus ever infect the host cell again. For millions of years, bacteria and viruses have been locked in an evolutionary arms race. Viruses evolving new mechanisms to evade CRISPR, and CRISPR evolving to find different ways of preventing re-infection. CRISPR-Cas13a is one such permutation. While other CRISPRs like Cas9 scan genes for viral DNA, Cas13a looks for viral RNA in the cytoplasm.
Pubmed Description 5xwp
Cas13a, a type VI-A CRISPR-Cas RNA-guided RNA ribonuclease, degrades invasive RNAs targeted by CRISPR RNA (crRNA) and has potential applications in RNA technology. To understand how Cas13a is activated to cleave RNA, we have determined the crystal structure of Leptotrichia buccalis (Lbu) Cas13a bound to crRNA and its target RNA, as well as the cryo-EM structure of the LbuCas13a-crRNA complex. The crRNA-target RNA duplex binds in a positively charged central channel of the nuclease (NUC) lobe, and Cas13a protein and crRNA undergo a significant conformational change upon target RNA binding. The guide-target RNA duplex formation triggers HEPN1 domain to move toward HEPN2 domain, activating the HEPN catalytic site of Cas13a protein, which subsequently cleaves both single-stranded target and collateral RNAs in a non-specific manner. These findings reveal how Cas13a of type VI CRISPR-Cas systems defend against RNA phages and set the stage for its development as a tool for RNA manipulation.
Dataset Deepdive
To understand the protein dynamics of CRISPR Cas13a described above, I have chosen a primary color scheme to showcase Cas13a’s atomic temperature by tinting cyan from dark to light according to it’s cooler and hotter regions. Like our other CRISPR models, Guide RNA is dark red and Target RNA is dark yellow.
As we go deeper into the data, I’ve revealed its ribbon backbone (cyan) and visualized all its 1154 residues as wires. Inside each twisting chain of nucleotides, I’ve colored each base (adenine yellow, cytosine red, guanine green, uracil cyan). Indicated in the pubmed abstract, two important domains (HEPN1 and 2) come together inside the positively charged central core to perform the cutting action on RNA. Here, I’ve colored these resides a darker blue color and assigned a stick representation to those atoms. Once the domains together, its catalytic site is activated (Leu771 colored orange) and subsequently cleaves each strand of RNA.
Model Description
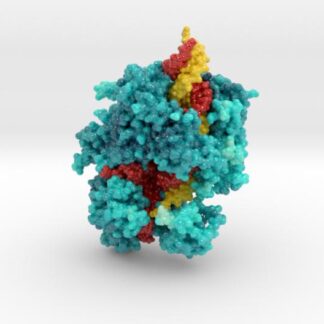
This is a 3D print of CRISPR Cas13a printed in full-color sandstone. Cas13a is colored turquoise and tinted according to the protein’s bFactor, or atomic temperature. The darker regions of Cas13a are the coolest, and the brightest the warmest. Colored yellow is the target RNA and red is the Guide RNA.
3D Print CRISPR-Cas13a
Custom 3D Print Request
Request a custom 3D printed protein model. Send us the protein name and PDB ID of the protein you’re interested in printing and we’ll get back to you with a feasibility analysis and estimate for printing.
[ninja_form id=6]