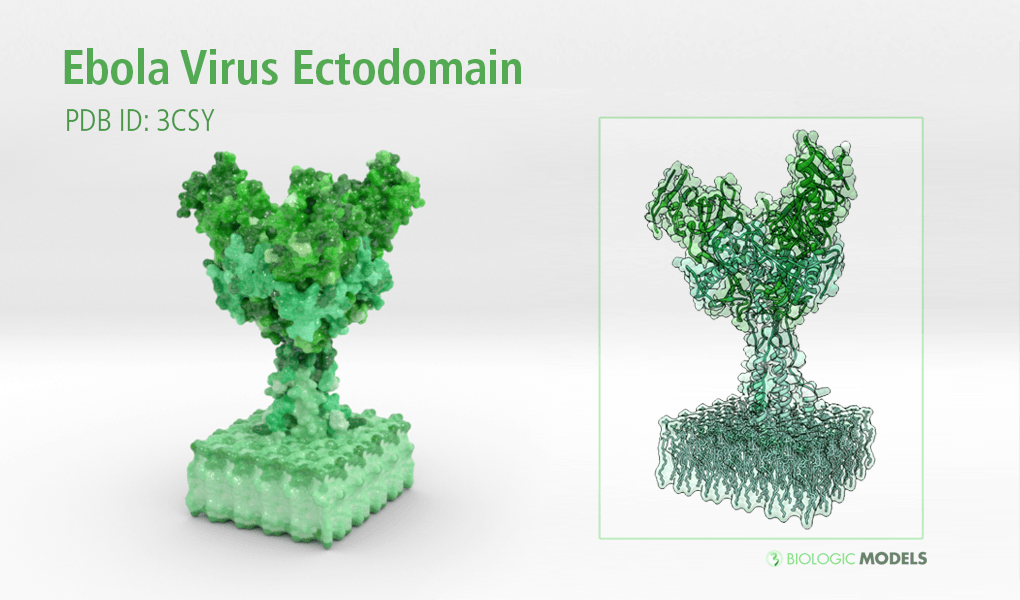
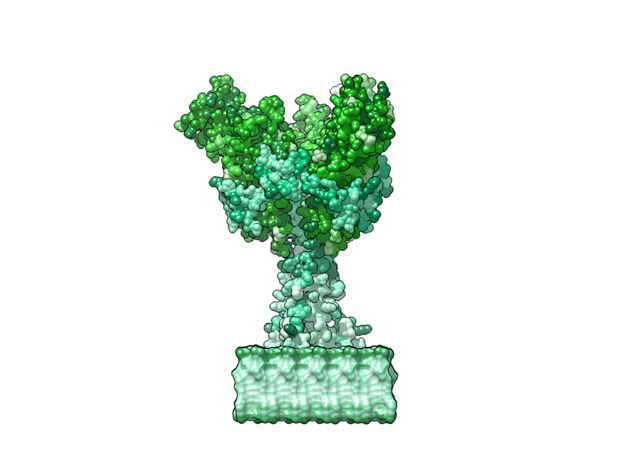
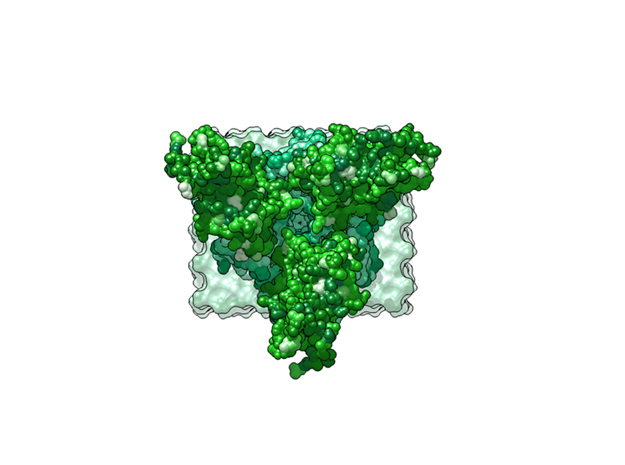
Ebola Virus Ectodomain
In this 3D exploration of the Ebola Virus Glycoprotein, we will investigate different surface properties and binding sites through 3D printed models and animation.
Protein Description
Ebola virus (EBOV) is a highly virulent pathogen capable of causing a severe hemorrhagic fever with 50–90% lethality. The EBOV glycoprotein (GP) is the only virally expressed protein on the virion surface and is critical for attachment to host cells and catalysis of membrane fusion. Hence, the EBOV GP is a critical component of vaccines as well as a target of neutralizing antibodies and inhibitors of attachment and fusion. The crystal structure of the Zaire ebolavirus GP in its trimeric, prefusion conformation (3 GP1 plus 3 GP2) in complex with a neutralizing antibody fragment, derived from a human survivor of the 1995 Kikwit outbreak, was recently determined.
This is the first near-complete structure of any filovirus glycoprotein. The overall molecular architecture of the Zaire ebolavirus GP and its role in viral entry and membrane fusion are discussed in this article.” — NCBI Citation
3D Animation
The ebola virus has a natural enemy, the human immune system. Those lucky enough to survey this deadly virus produce an antibody which identifies the virus for destruction. The above animation shows these antibodies attached to the ectodomains of the virus.
Structural Mechanisms of Nucleosome Recognition by Linker Histones
“The EBOV envelope GP directly mediates binding of the virion to the host cell. A number of cellular factors, including DC-SIGN/L-SIGN [68,69], LSECtin [70,71], hMGL [72], β-integrins [73] and Tyro3 family receptors [74], have been implicated as attachment factors, however, none of these proteins individually are necessary and sufficient for viral entry. Hence, a critical cell-surface receptor responsible for EBOV attachment has yet to be identified, despite significant effort. It appears that EBOV enters through a receptor-mediated endocytotic mechanism, but it is still unclear whether clathrin-, caveolae- or cholesterol-dependent processes are used. Recombinant systems using pseudotyped retro-virus particles with ZEBOV GP and live virus studies using chemical inhibitors of clathrin- and caveolae-mediated endocytosis point to the use of caveolae and clathrin in EBOV entry [75,76]. However, cells lacking in caveolae can still be infected with EBOV [77].” – NCBI Citation
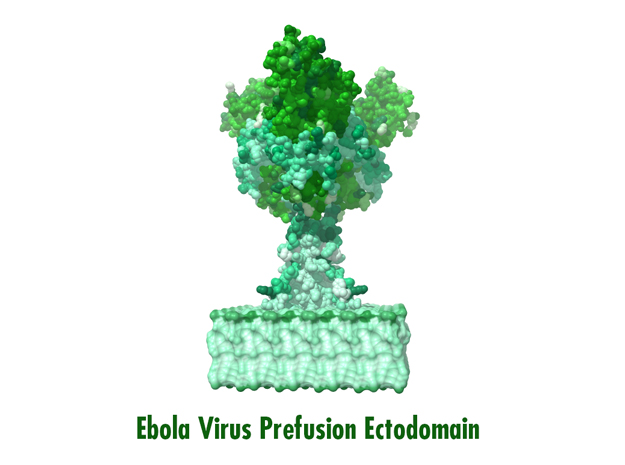
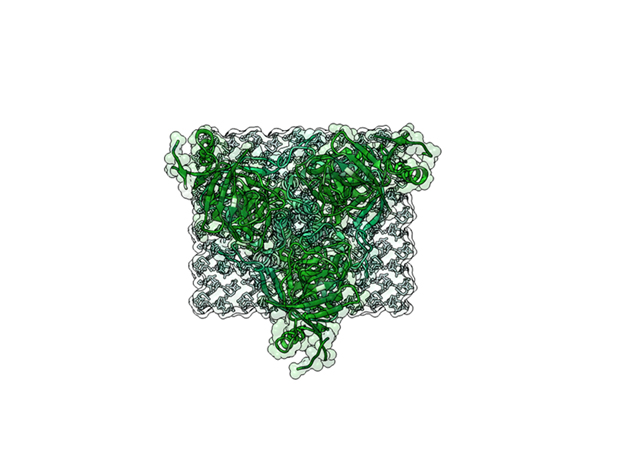
Model Description
This is a 3D print of x-ray crystallography datasets PDB IDs: 3CSY, 2EBO The protein surface color coding is defined by the glycoprotein’s Hydrophobicity. To delineate the different chains, we have colored the GP2-Glycoprotein-plasma membrane shades of green. The model can be printed in a variety of materials and sizes.
Purchase Model
Custom 3D Print Request
Request a custom 3D printed protein model. Send us the protein name and PDB ID of the protein you’re interested in printing and we’ll get back to you with a feasibility analysis and estimate for printing.
[ninja_form id=6]